
|
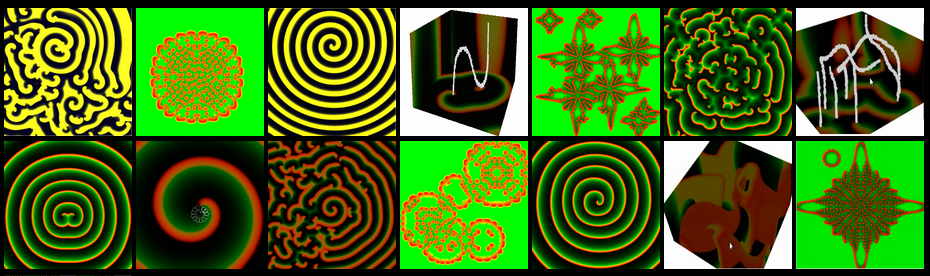
We present here a set of interactive programs to study and analyze several models of excitable media in tissue. As the waves they produce propagate through the media, the models exhibit complex spatiotemporal dynamics that cannot be appreciated from an analysis of the underlying equations or even verbal descriptions. Here, we allow users to perform in real time simulations of these models and to watch the patterns develop and change over time as the simulated dynamical waves propagate. The parameters governing the model's behavior can be changed on the fly to alter the dynamics. In addition, users can apply perturbations and periodic pacing, that change the patterns locally an globaly and watch the response. One of the main advantages of these programs is that the models are implemented using WebGL, which allows the simulations to be run over the Internet, independent of computer architecture and operating system. WebGL utilizes available hardware, including graphics cards, to improve performance, thereby allowing simulations to run as a parallel implementation on the computer's GPU and to respond rapidly to user interactions. In fact, some smart phones are capable of running these simulations. Our implementations also feature very large domains, (256x256,512x512 and 1024x1024) in 2d and one 3D, that provide sufficient space for complex and interesting patterns to form and evolve. We have implementated three generic models of excitable media, the FitzHugh-Nagumo model, the Barkley model (which is a simplification of the FitzHugh-Nagumo model), and the Gray-Scott model, which among other things includes an unusual backfiring feature. In addition, we have implemented two models specific to excitable cardiac electrophysiology that develop complex spatiotemporal patterns, the Karma model and the BuenoOrovio–Cherry–Fenton model. Altogether, these models provide a fascinating portrait of complex spatiotemporal dynamics in excitable media. WebGL ProgramsNote: You will need Google Chrome or Mozilla to play the WebGL programs. With mozilla in some cases it may be needed to give permision for webGL to run, which is very simple as shown here. FitzHugh-Nagumo model [1]∂U/∂t = F(U, V) = Du∇2U + (U(1 - U)(U - a) - V),∂V/∂t = H(U, V) = (bU - V - δ)ε
Barkley model [2]∂U/∂t = Du∇2U + U(1 - U)(U - Uth ) ,∂V/∂t = (U - V)ε , Uth = (V - b)/a .
Gray-Scott model [3]∂U/∂t = Du∇2U + (U2V - aU)/ε∂V/∂t = Dv∇2V - U2V +(b-cV)
Karma model [4]dE/dt = F(E, n) = [A - (n/nb)M][1 - tanh(E - 3)]E2/2 - E,dn/dt = H(E, n) = ε [Θ(E - 1) - n]. where A = 1.5415.
BuenoOrovio-Cherry-Fenton model [5]du/dt = -(Jfi + Jso + Jsi ) ,dv/dt = [1 - H(u - Vv )](v∞ - v)/τv - - H(u - Vv )v/τv , dw/dt = [1 - H(u - Vw )](w∞ - w)/τw- - H(u - Vw )w/τw , ds/dt = ((1 + tanh[ks (u - Us )])/2 - s)/τs ,
Main Bibliography: [1] FitzHugh-Nagumo model in Scholarpedia [2]Dwight Barkley A model for fast computer simulation of waves in excitable media Physica D 49 (1991) 61-70. [3] P. Gray and S.K. Scott, Autocatalytic reactions in the isothermal, continuous stirred tank reactor: Oscillations and instabilities in the system A+ 2B --> 3B; B --> C Chem. Engng. Sci. 39, 1087 (1984). [4] Alain Karma Spiral Breakup in Model Equations of Action Potential Propagation in Cardiac Tissue Phys.Rev.Lett. 71, 1103 (1993) [5] Bueno-Orovio A., Cherry EM, Fenton FH. Minimal Model for Human Ventricular Action Potential in Tissue. Journal of Theoretical Biology 2008; 253 544-560. Supplemental Bibliography: [6]A. T. Winfree, Varieties of spiral wave behavior in excitable media , Chaos 1, 303–333 1991. [7] V. N. Biktashev, A. V. Holden, and H. Zhang, ‘‘Tension of organizing filaments of scoll waves,’’ Philos. Trans. R. Soc. London, Ser. A 347, 611–630 1994. [8] M. R. Guevara, G. Ward, A. Shrier, and L. Glass, ‘‘Electrical alternans and period doubling bifurcations,’’ Comput. Cardiol. 167–170 1984. [9] Watanabe M, Fenton F, Evans S, Hastings H, Karma A. Mechanisms for Discordant Alternans. Journal of Cardiovascular Electrophysiology 2001; 12: 196-206. [10] Bartocci E, et al. “Teaching cardiac electrophysiology modeling to undergraduate students: Lab exercises and GPU programming for the study of arrhythmias and spiral wave dynamics” Advances in Physiology Education, 35: 427-437. 2011. [11] Models of Cardiac Cell in Scholarpedia. By: Evgeny Demidov, demidov@ipm.sci-nnov.ru Elizabeth M. Cherry, excsma@rit.edu and Flavio H. Fenton, flavio.fenton@physics.gatech.edu  Part of this work was performed under NSF grant # 1341190
Collaborative Research: Dynamical Systems Program.
Part of this work was performed under NSF grant # 1341190
Collaborative Research: Dynamical Systems Program.
|